SERVICES>Examples of tests>Real-time PCR
|
Real-time PCR
These are images of a DNA detection technique called real-time PCR. With this technique a gel is not needed and the DNA being copied is detected during the PCR process by a laser detecting a fluorescent dye that binds to DNA. Real-time PCR is faster and more sensitive than detecting by gels, but the reagents are more costly. Each curve or line is a different sample. The x-axis is the number of cycles that the DNA has been copied. The y-axis is the amount of fluorescence detected. As the number of cycles increases (more copying) the fluorescence increases (more DNA). If one curve is more to the left of the others, it started with more DNA in the tube (amplifies sooner). Likewise, if a curve is more to the right of the others, it started with less DNA in the tube. There are many ways to use fluorescence to detect the DNA. Each method produces amplification curves similar to the example shown in the figure. Two methods are the most common: SYBR Green - a fluorescent dye that binds to double stranded DNA in a quantitative way. It binds to any double stranded DNA, and thus is non specific for a particular DNA. When this dye is used it is important to verify the specificity of the PCR amplification by either a melt curve after the PCR run, or by running on a gel, or both. Fluorescent probes - short fragments of single stranded DNA, designed by the user and synthesized by a company, that bind to a particular exact sequence of DNA. The probe is synthesized with both a fluorescent molecule and its quencher attached so that it cannot fluoresce until it is time. The probe is designed to attach to the region of DNA that is being copied. When the DNA is copied, the probe is chopped up and the fluorescent molecule is released and can then fluoresce and be detected by the instrument. If designed properly, probes bind to only one location in the DNA being tested and nowhere else. These probes are highly specific. They are commonly called TaqMan probes. |
Testing of corn DNAs for GM sequences
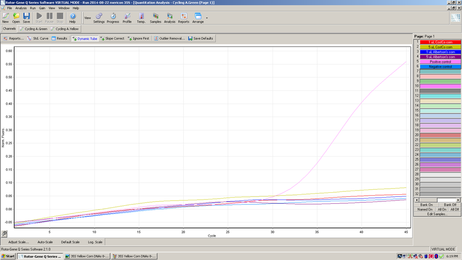
This graph represents amplification using a green fluorescent probe to detect the 35S gene promoter commonly used in GM plants.
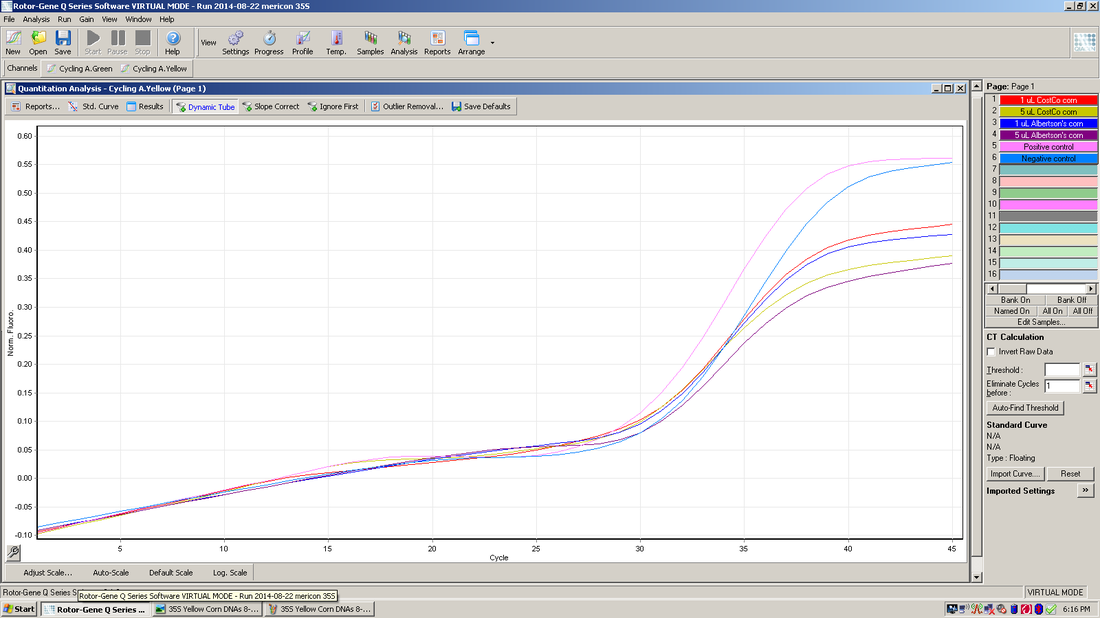
This graph represents a yellow fluorescent probe to detect an internal control that is included in each of the samples.
The experiment above is the result of real-time PCR using two fluorescent probes in each of the six samples. Four of the samples are corn DNAs, and the other two are a positive control and a negative control. There is a separate window or result for each color of probe used. The top graph shows the results for the green probe that was designed to bind to the 35S promoter. None of the corn DNAs are positive for this promoter. The only sample to amplify with this probe is the positive control supplied with the kit reagents. The bottom graph shows the results for the yellow probe that is an internal control supplied with the kit. The internal control is added to each of the samples and in this experiment it amplified properly in all 4 corn DNAs and both controls, although the four corn DNAs did not reach the same level (height) as the two control samples. The internal control is also designed to determine whether there are any inhibitors to the PCR reaction in the sample being tested. In this experiment it appears there is a slight inhibitor in each of the four corn DNAs. If there would be a complete inhibition of the PCR reaction, this result could appear as a false negative. |